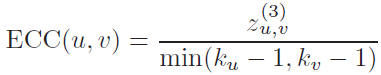EGC - Edge clustering coefficient and Gene ontology information’s Combination method
Definition
Gene ontology (GO) information is adopted as a
measure to evaluate the reliability of the edges in PPI network and a new algorithm
EGC is proposed to identify essential proteins by integrating the topological features
of the PPI network and the information of GO.
The essentiality of each protein u in PPI network is determined by: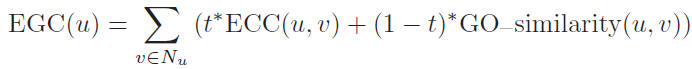 Here, t is a proportionality parameter which takes value in the range of 0 to 1, Nu
is the set which contains all the neighbors of u.
Here, t is a proportionality parameter which takes value in the range of 0 to 1, Nu
is the set which contains all the neighbors of u.
GO similarity
The GO similarity between protein u and protein v is
Edge Clustering Coefficient
The edge clustering coefficient of an edge (u, v), connecting node u and node v, can be defined by the following formula:
The essentiality of each protein u in PPI network is determined by:
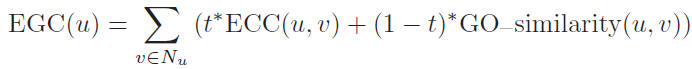
GO similarity
The GO similarity between protein u and protein v is
- GO–similarity(u, v) = GO(u) ∩ GO(v)
Edge Clustering Coefficient
The edge clustering coefficient of an edge (u, v), connecting node u and node v, can be defined by the following formula:
Requirements
Undirected graph G=(V,E), GO database, parameter t and k.
Software
References
- LUO, J. & ZHANG, N. 2014. Prediction of essential proteins based on edge clustering coefficient and gene ontology information. Journal of Biological Systems, 22, 339-351.
DOI: 10.1142/S0218339014500119